Dr Liam Cox, Project Lead
University of Birmingham
We need to be better at drug discovery
To respond rapidly to emerging medical emergencies, we must become more efficient at developing drugs, ideally by reducing the attrition rates that blight the drug discovery and development process. One approach to achieving this goal is to populate compound screening libraries with more structurally diverse and higher quality molecules. This aim has been the focus of project iDesign.
Identifying the right starting point
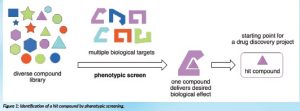
A drug molecule typically acts upon a specific biological target. The drug prevents the target from playing its role in a disease or condition, leading to a therapeutic treatment. In many cases, a defined biological target is not available; in others, there is a desire to identify novel targets, for example, if existing drugs, acting upon well-established targets, are losing their efficacy; this is the case for many antimicrobials owing to the emergence of bacterial resistance. In these scenarios, an attractive starting point for a drug discovery project is to perform a phenotypic screen (Figure 1).
Say, for example, we want to develop a new treatment for tuberculosis; if we expose the causative agent of the disease, the bacterium Mycobacterium tuberculosis, to a molecule that results in the death of the bacterium, we have identified a molecule that has the desired antimicrobial effect. This so-called ‘hit’ molecule now provides the starting point for developing an anti-tuberculosis agent. Unfortunately, the likelihood of identifying a molecule that has a desired effect is very low. To improve our chances, phenotypic screens are typically performed using compound libraries that can contain many thousands of different molecules. To further improve our chances of identifying a hit, compounds in a screening library need to be structurally diverse; this allows us to ‘sample’ a large area of so-called chemical space and increase the probability that one molecule in our library finds a suitable target amongst the many thousands that are available.
Having identified a hit molecule from a phenotypic screen, we next need to identify its biological target. If the target proves to be ‘druggable’, structural modifications to the starting hit introduce the physicochemical properties that are needed to transform the hit into a drug. This process is long and complex, and there are many reasons why a project might fail. However, we can improve our chances of success if our starting hit already possesses a good safety profile and structural and physicochemical properties that are conducive to further development. In this way, we can not only increase the rate of development but also the likelihood that the project reaches a successful outcome, namely a new therapeutic treatment for use in the clinic.
iDesign’s approach to smart library synthesis
Project iDesign sought to address the challenges of compound library design through a collaboration between researchers from academia and industry. The project started in 2018 with the recruitment of six early-career researchers who enrolled as PhD students at the University of Birmingham, UK. All six spent the first half of their three-year PhD projects at Birmingham, completing an expansive training programme while also undertaking the first phase of their PhD research projects, synthesising a diverse range of molecular scaffolds using chemistry developed by their academic supervisors at Birmingham. Critically, each scaffold was validated first in silico. This de-risking exercise ensured that only structurally novel compounds predicted to exhibit favourable physicochemical properties and safety profiles were prioritised for synthesis.
In January 2020, all six researchers moved into industry to complete the second phase of their PhD projects, four to Symeres in The Netherlands and two to AnalytiCon Discovery in Germany. Here they explored the chemical reactivity of functionality that had been purposely embedded in their scaffolds to establish methods for attaching a diverse array of appendages, using transformations that have a demonstrable record of successful application in drug discovery and development. The PhD researchers’ industrial supervisors now guided research efforts and trained the researchers in the parallel synthesis methods needed to efficiently design, prepare and purify compound libraries.
Library design saw the researchers again using in silico methods, this time to enumerate compound libraries from each of their validated molecular scaffolds. Workflows were developed to ensure enumerated compound libraries interrogated a maximal region of drug-like chemical space, considering shape, novelty and key physicochemical property descriptors for small-molecule drug development. Workflows incorporated operations that filtered out potential toxicophores and sub-structures (so-called PAINS) that are known to interact non-specifically with a broad range of biological targets. Given their size, in silico clustering methods were then used to reduce these virtual compound libraries to more synthetically tractable libraries of around 100–150 compounds while still retaining the diversity of the enumerated libraries.
While in silico methods had provided valuable de-risking tools, representative compounds from the researchers’ synthesised libraries were assessed for hERG activity. The human Ether-á-go-go-Related Gene (hERG) codes a protein that is a key component of an important potassium ion channel in the heart. This ion channel is sensitive to drug binding, which can result in decreased channel function, leading to adverse cardiac effects (arrhythmia), and in the worst cases, can lead to death. Irrecoverable inhibition of the hERG protein can therefore lead to the cessation of a drug discovery project. hERG screening was undertaken by our project partner, ApconiX. Some compounds exhibited levels of hERG inhibition that would likely lead to a safety flag in a drug discovery project; however, inhibition was typically associated with the scaffold appendages. This observation is significant; it demonstrates that the scaffolds themselves are not inherently responsible for hERG inhibition and highlights how modification of appendages would provide a strategy for addressing hERG issues in a future drug discovery project.
In total, the iDesign project delivered a new physical collection of 1 093 compounds, which have been added to the commercially available collections maintained by Symeres and AnalytiCon Discovery. A sub-set of compounds from these collections was transferred to the University of Birmingham’s own compound collection.
Attractive starting points for drug discovery
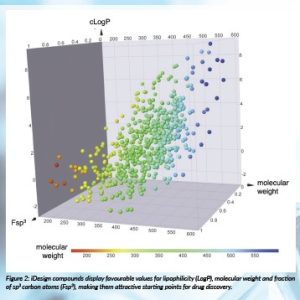
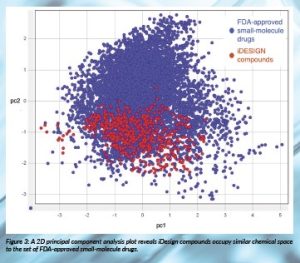
iDesign’s library compounds were designed from the outset to display favourable physicochemical properties for drug discovery (Figure 2). They occupy a similar chemical space to small-molecule drugs already approved by the US Food and Drug Administration (FDA) (Figure 3) but exhibit high levels of dissimilarity from these drugs, confirming the structural novelty of the iDesign collection. Compounds have been experimentally validated, and the scaffolds shown to display favourable stability, solubility and safety profiles, which should render hits arising from screening our libraries attractive starting points for drug discovery.
Early screening results demonstrate the value of the iDesign compound library
Initial screening activities have been undertaken by the University of Birmingham’s Drug Discovery Facility. A representative set of 565 compounds from the six researchers’ compound libraries, were screened against Mycobacteria and a set of ESKAPE pathogens, which are bacteria for which there is a particular pressing need to develop new antimicrobial agents. Hit compounds derived from three different scaffolds showed significant activity against Mycobacterium smegmatis (model organism for M. tuberculosis, the causative agent of tuberculosis) and Gram-positive bacteria, notably Enterococcus faecium E745, a vancomycin-resistant strain, listed as a high-priority target on the World Health Organisation’s priority pathogen list. Future work will focus on validating these exciting early results and undertaking target identification studies.
In silico library enumeration employed reactions that had been experimentally validated on the researchers’ scaffolds. The resulting enumerated libraries, therefore, provided a set of 3164 synthetically tractable compounds; these have been deposited in the University of Birmingham’s virtual compound collection. A recent screen of this collection identified a series of compounds as putative inhibitors of a protein-protein interaction that researchers in the Centre for Liver and Gastroenterology Research at Birmingham have identified as a target for developing a treatment for acute alcoholic hepatitis. The compounds have now been synthesised, and we await the results of physical screening to validate these promising in silico studies.
Knowledge transfer
The iDesign project has provided a great opportunity for knowledge transfer between all partners. During their industrial placements, the researchers encouraged the take up of types of chemistry that have not (yet) received widespread application within industry. For the Birmingham team, working closely with industrial partners provided invaluable insight into practical library design and helped to formulate industry-standard criteria for developing the University’s own compound collection. The iDesign project has also catalysed a significant drive to develop drug discovery activities across the University, with the establishment of the Birmingham Drug Discovery Hub, which brings together researchers from across the University and industry to develop drug discovery programmes. The hub promises to be a long-lasting legacy for this EU-funded training network.
Project name
iDesign
Project summary
Project iDesign’s primary research objective is to deliver better starting points for early-stage drug discovery. To this end, we have synthesised compound libraries comprising molecules that are demonstrably novel, structurally and functionally diverse, and lead-like in character. Designed with safety considerations from the outset, our libraries interrogate underexplored regions of chemical space and therefore complement existing diversity sets.
Project partners
Project iDesign is a European Industrial Doctorate Innovative Training Programme involving The University of Birmingham (UK) as the Academic Beneficiary and two Industrial Beneficiaries, Symeres (The Netherlands) and AnalytiCon Discovery (Germany), with Project Partner ApconiX (UK) bringing their expertise in drug safety to the project.
Project lead profile
Dr Liam Cox is a synthetic organic chemist. He joined The University of Birmingham in 1999 and is currently a Reader in Biological Organic Chemistry. His research increasingly lies at the interface of chemistry and the life and medical sciences, where his team apply their synthesis skills to solving biological problems and early-stage drug discovery.
Project contact
Dr Liam Cox, Project Lead
University of Birmingham
Funding
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 765116.
Figures
Figure 1: Identification of a hit compound by phenotypic screening.
Figure 2: iDesign compounds display favourable values for lipophilicity (LogP), molecular weight and fraction of sp3 carbon atoms (Fsp3), making them attractive starting points for drug discovery.
Figure 3: A 2D principal component analysis plot reveals iDesign compounds occupy similar chemical space to the set of FDA-approved small-molecule drugs.