Nele Vandersickel
What does it mean to advance science? Should we try to publish as many papers as possible? Or should we try to impact the lives of real people?
1. Impactful, reproducible and sustainable science
A quick Google search tells me that there are about 30 000 scientific journals, which publish about two million scientific papers each year. This is a huge amount of information, which has grown exponentially in the last decades (Fire and Guestrin, 2019). In academia, success is sometimes measured by the number of papers a researcher publishes. However, when a measure becomes a target, it ceases to be a good measure, which was stated by Goodhart (Strathern, 1997). Therefore, we as scientists need to rethink how we evaluate good science. A higher number of published papers than ever before are required to obtain grants, positions, promotions and status. This could lead to fractionation (‘salami slicing’ data sets to the smallest publishable unit), surreptitiously re-using data in multiple publications, duplicating papers, publishing results that are preliminary or incomplete, under-emphasising limitations, making exaggerated claims, data fabrication, falsification and plagiarism. Therefore, we can ask, how can an academic research team make an impact in this enormous mass of published science?
The replication crisis shows that many results claimed in scientific/medical papers are not reproducible (Baker, 2016), suggesting that many of the two million papers should not have been published in their current form. This creates a lot of inefficiencies: people are reading papers that contain flaws as they cannot be reproduced, creating information that is simply not true. Therefore, in addition to all the time wasted in reading non-reproducible papers, researchers also do not know which research is reproducible and which is not. On top, researchers who publish non-reproducible papers should have spent more time creating scientific content which is reproducible, as writing non-reproducible papers is also a waste of their time. While it is clear that we scientists should change the system of evaluation, it also raises the question, how can we create science that is reproducible?
A last important factor in science is what happens with your work when you finish a project. More and more research relies on custom software, which often is lost or no longer maintained when the researcher finishes their work in case there is no follow up. If another scientist wants to re-use a certain part of novel techniques, one needs to rewrite code to be able to use certain scientific methods. This, again, is very inefficient, and many hours could have been saved if we started to use better practices. Therefore, how can we create (software) science that is sustainable?
My ERC starting grant allowed me to think about these three important questions raised in the previous section. I will share how I address them in section three while first explaining the scientific project in detail in section two.
2. Directed Graph Mapping (DGM)
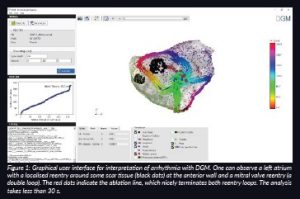
We have developed a software package called Directed Graph Mapping (DGM), which can automatically determine the origin of the arrhythmia during an ablation procedure. Let us explain this in a bit more detail: the heart consists of four parts, two upper chambers called the atria, and two lower chambers, the ventricles. The contraction of the heart is initiated by an electrical wave that periodically starts at the sinus node node in the right atrium. From there, the electrical wave spreads through the whole right and left atrium, and then further to both ventricles. This electrical wave allows efficient circulation of the blood. We will focus on one type of arrhythmia, named atrial tachycardia (AT), as DGM is currently optimised for this type of arrhythmia. One of the main driving mechanisms for AT is a regular anatomical reentry, which occurs when the electrical wave starts to rotate around an obstacle, e.g. around a valve, a vein or scar tissue. Anatomical reentry can be easily compared to a Mexican wave, which in principle does not stop by itself. Similarly, anatomical reentry also sustains itself and is usually very fast, superseding the normal rhythm of the heart of the sinoatrial node. One way to cure the arrhythmia is by burning a part of the heart (ablation), which can be compared to building a wall in the Mexican wave, so it would stop. During an ablation, the cardiac electrophysiologist (EP) will measure the electrical activity of the heart: the local activation times (LAT = the exact moment the electrical waves passed a given location). In the current clinical workflow, these measurements are converted to a coloured map of the heart by a mapping system, which then needs to be manually interpreted by the EP to find the location of the arrhythmia. This can lead to different diagnoses depending on the interpretation by the EP, which in turn can lead to the placement of unnecessary ablation lines. Not only can these unnecessary ablation lines reduce the function of the heart, but they can also be pro-arrhythmic.
DGM takes as input the same measurements of the heart performed by the EP (the LATs and their coordinates) and converts these measurements into a virtual directed network that represents the excitation of the heart. We were the first group in the world to take this interdisciplinary approach to cardiac arrhythmia. Network theory is often used to find the underlying structure in physical, biological, social and information systems. For example, network theory is the basis of Google’s search engine technology. Network theory is increasingly being applied to the brain (Giusti, Ghrist and Bassett, 2016; Bullmore and Sporns, 2009) to uncover fundamental architectural principles (Bassett and Bullmore, 2006; Bullmore and Bassett, 2011; Park and Friston, 2013) and has led to the development of novel diagnostic biomarkers for Alzheimer’s, multiple sclerosis, traumatic brain injury and epilepsy (Stam, 2014), to name a few. By converting an arrhythmia to a directed network, we can easily detect cycles or closed loops in this network using algorithms from different research fields. These cycles represent the anatomical reentry. By optimising our algorithms, DGM automatically finds the correct location of the arrhythmia, independent of any manual interpretation. This makes DGM operator independent. In Figure 1, we show a clinical case of a double loop reentry which was analysed by DGM.
3. Creating true impact with a reproducible and sustainable software package DGM
3.1 Impact
With our scientific developments, our group is determined to make an impact on the medical world. Currently, DGM performs better than the conventional mapping systems, as we also know how to treat and detect double loop reentries (two simultaneous regular anatomical reentries). DGM is the first software package worldwide which does not need any manual interpretation of a given mapped arrhythmia. We spent over five years developing this technology from the initial idea to a full-scale software package (Vandersickel et al., Van Nieuwenhuyse et al., 2017). We recently (October 2021) released the first version of our software under an academic licence which can be obtained via our website www.dgmapping.com. We are now working to incorporate our novel findings concerning double loop ATs, and we hope to gain interest of mapping companies in the year 2022. If such a company implemented DGM, we would be able to help patients by providing a robust, operator-independent diagnosis. In time, we will also provide the optimal ablation lines for each given case. Moreover, we have a proof-of-concept that DGM can uncover suppressed loops which will take over the arrhythmia after conventional ablation. Currently, none of the existing mapping systems know how to find these loops. Therefore, it requires a second map after ablation of the first AT, or even a redo in a later procedure. Consequently, if we can further confirm our hypothesis, only a single map will be required, allowing significantly shorter mapping times (as only a single map is required) and a possible reduction in redo operations, thereby significantly reducing the burden on patients.
3.2 Reproducibility
We have spent an ample amount of time making DGM widely applicable. We have incorporated the formats of the two main mapping systems (Boston Scientific and Biosense Webster), and we have tested DGM on over 150 cases of AT ourselves. Recently, we have released the software for other groups to test as well. We have spent a large amount of time creating a stable easy-to-use package so many different groups and EPs can independently test DGM. As medical data is protected, easy exchange of data is not possible. We hope that by releasing the software, we will obtain valuable feedback to improve our software. Although this work does not create new publications, it is crucial to make an impact in the scientific world. However, this is currently not incorporated in the evaluation of good science despite being crucial.
3.3 Software sustainability
In addition, we have also spent a huge amount of time in making the source code of the software well maintained, well documented and modular so it can be easily extended and to allow easy testing of other approaches in a specific part of the entire flow without having to rework large parts of the codebase. Although this does not immediately translate into scientific output, it makes the scientific process much more efficient. A description of the software is accepted in the journal Medical and Biological Engineering and Computing. Having modular software also helps to extend the research to other arrhythmias, as next to AT, we are also performing research on atrial fibrillation (AF) (Van Nieuwenhuyse, 2021) (plus new studies), on Torsade de Pointes (Vandersickel, 2017) (plus a submitted paper), ventricular fibrillation (under investigation) and ventricular tachycardia (submitted paper).
4. Future perspectives
As our goal is to bring our diagnostic tool to the patient, it is important that multiple EPs test our software to confirm if they find our tool a valuable addition to the current mapping process. For this, we need to disseminate our research to a large group of EPs. If multiple EPs would ask their current mapping company to add DGM, then the chances increase for DGM to enter the clinical workflow.
So far, DGM has only been extensively tested on AT. Despite it being considered an ‘easy’ arrhythmia, we have found many novel scientific questions in AT. However, we believe many new challenges lie ahead. The power of DGM is that it is independent of the chosen measuring system of the arrhythmia. It can analyse any type of dataset from which we can extract LATs and spatial coordinates. The holy grail of arrhythmias is AF, for which the prevalence is rising steeply due to an ageing population. Although AF by itself is not lethal, it causes a sixfold increase in stroke risk, leads to anxiety and depression, and severely reduces quality of life. Currently, the treatment of certain types of AF remains suboptimal. The main reason is that the mechanisms of long standing AF is still not well understood. Leading groups have opposing views on whether AF is maintained by reentry, focal sources or by more complex combinations. The current methods to analyse AF have been shown to be sensitive to errors. Therefore, it is our hope that our new analysis technique will enhance understanding of the mechanism of AF and lead to novel methods to ablate this arrhythmia. As this arrhythmia is not regular, point by point measurements cannot be used for DGM, and we have to rely on non-contact mapping systems to get a picture of the whole arrhythmia. Alternatively, we can only analyse a small simultaneous mappable part of the atrium. A first computational study already revealed that DGM is superior to phase mapping for finding rotors in a particular non-contact dataset of a drifting rotor (Van Nieuwenhuyse, 2021).
References
Baker. M., (2016) ‘1,500 scientists lift the lid on reproducibility’ Nature, 533, pp. 452–454. doi: 10.1038/533452a.
Bassett, D.S. and Bullmore, E.T. (2006) ‘Small-world brain networks’, Neuroscientist, 12(6), pp. 512–23. doi: 10.1177/1073858406293182.
Bullmore, E.T and Sporns, O. (2009) ‘Complex brain networks: graph theoretical analysis of structural and functional systems’, Nature Reviews Neuroscience, 10(3), pp. 186–198. doi: 10.1038/nrn2575.
Bullmore, E.T. and Bassett, D.S. (2011) ‘Brain graphs: graphical models of the human brain connectome’, Annual Review of Clinical Psychology, 7, pp. 113–140. doi: 10.1146/annurev-clinpsy-040510-143934.
Fire, M. and Guestrin, C. (2019) ‘Over-optimization of academic publishing metrics: observing goodhart’s law in action’ GigaScience, 8(6). doi: 10.1093/gigascience/giz053.
Giusti, C., Ghrist, R. and Bassett, D.S (2016) ‘Two’s company, three (or more) is a simplex’, Journal of Computational Neuroscience, 41(1), pp. 1–14. doi: 10.1007/s10827-016-0608-6.
Park, H-J. and Friston, K. (2013) ‘Structural and functional brain networks: from connections to cognition’, Science (New York, N.Y.), 342, 1238411. doi: 10.1126/science.1238411.
Stam, C.J. (2014) ‘Modern network science of neurological disorders’, Nature Reviews Neuroscience, 15(10), pp. 683–695. doi: 10.1038/nrn3801.
Strathern, M. (1997) ‘’Improving ratings’: audit in the British University system’, European review, Cambridge University Press, 5(3), pp. 305–321. doi: 10.1002/(SICI)1234-981X(199707)5:3<305::AID-EURO184>3.0.CO;2-4.
Van Nieuwenhuyse, E., Mateu, L.M., Saiz, J., Panfilov, A. and Vandersickel, N. (2021) ‘Directed graph mapping is able to distinguish between the true and false rotors in a complex in-silico excitation pattern’, Europace, 23. doi: 10.1093/europace/euab116.544.
Vandersickel, N., Bossu, A., De Neve, J., Dunnink, A., Meijborg, V.M.F., van der Heyden, M.A.G., Beekman, J.D.M., De Bakker, J.M.T., Vos, M.A. and Panfilov. A.V. (2017) ‘Short-Lasting Episodes of Torsade de Pointes in the Chronic Atrioventricular Block Dog Model Have a Focal Mechanism, While Longer-Lasting Episodes Are Maintained by Re-Entry’, JACC: Clinical Electrophysiology, 3(13), pp. 1565–1576. doi: 10.1016/j.jacep.2017.06.016.