Claudia Bartoli1,2 and Anne-Yvonne Guillerm-Erckelboudt2
1LIPME, INRAE, CNRS, Université de Toulouse, Castanet-Tolosan, France
2IGEPP, INRAE, Institut Agro, Université de Rennes, 35650 Le Rheu, France
The evolutionary processes underlying interactions between hosts and their associated microbes are a black box on which biologists have long attempted to shed light. A limiting factor has been the knowledge of the precise molecular events, which are now increasingly possible to elucidate with the advent of next-generation sequencing technologies. Both big data and the artificial intelligence era have revolutionised evolutionary studies by sequencing thousands of genomes, allowing complex analyses to trace genomic changes.
However, fundamental questions about coevolution among interkingdom organisms remain largely unanswered, as analytical approaches alone—lacking biological experimentation in ecologically realistic contexts—are insufficient to fully capture coevolution dynamics. By integrating cutting-edge analytical technologies with in vivo experimental evolution, we can transition evolutionary science from a predictive framework to a functional discipline, enabling the observation of evolutionary changes in real time.
The key challenge now facing evolutionary biology is the identification of trajectories that simultaneously shape the evolution of hosts and their associated microbial communities. In this context, HoloE2Plant aims to validate the holobiont theory (considering the host and microbiome genomes as a unique entity on which the evolution acts) by exploring the coevolutionary dynamics of hosts and their microbiomes, providing unprecedented insights into the intricate interplay between these intimately connected biological entities.
To tackle this ambitious challenge, we developed an experimental coevolution approach using fast-cycling Brassica rapa (B. rapa) plants and synthetic microbial communities (SynComs) assembled from an extensive collection of bacterial and fungal isolates. HoloE2Plant contributes to validating the holobiont concept by providing unique methodologies to track rapid evolutionary changes within holobionts. These theoretical advancements will lay the foundation for future applied projects focused on designing microbial consortia with enhanced biocontrol properties.
Unveiling real-time evolution between plants and microbes requires building synthetic microbiomes from nature
Studying real-time evolutionary interactions between plants and microbes requires the reconstruction of artificial microbiomes that accurately reflect the microbial diversity found in natural habitats. Regrettably, agricultural soils often lack this diversity due to decades of intensive soil management and pesticide use, which have significantly depleted microbial communities inhabiting agricultural soils. This is particularly true for fungal communities, which are extremely sensitive to soil labour. To address this limitation, and establish microbial collections that can restore agricultural soil diversity, it is crucial to explore and isolate microbes from undisturbed natural habitats where soil use has minimal impact.
A major challenge lies in the fact that a significant fraction of fungal and bacterial communities associated with plants remain difficult to culture using traditional methods developed by Pasteur. Very often, few culture media are used in laboratories to isolate microbes, and these media do not reflect the natural substrate where microbes evolve. To capture a more comprehensive representation of natural microbial diversity, it is essential to employ a wide range of culturing media and advanced methodologies that encompass classic microbiological approaches and mimicking natural habitats.
In HoloE2Plant, we went back to nature to collect fungi and bacteria associated with wild B. rapa plants (Figure 1) growing in non-agricultural habitats. These habitats were selected because they display a large diversity of neighbouring plant species. By adopting an exhaustive culturomics approach— combining diverse microbial isolation techniques with high-throughput sequencing—we built an exceptional fungal and bacterial collection, showcasing remarkable species diversity (Figure 1). This incredible diversity was captured because we developed specific media mimicking soil habitats and adopted several culturing conditions. We prioritised unique fungal and bacterial species for in-depth characterisation, focusing on their ability to prevent attacks against the soil-borne fungal pathogen Rhizoctonia solani. Each strain was also phenotyped for its capacity to degrade a wide range of carbon sources using a high-throughput automated system. This analysis is crucial for avoiding carbon source overlaps among microbial species when constructing SynComs. Minimising competition for nutrients ensures that the microbial communities can effectively colonise the plant host and persist throughout its lifecycle.
Using insights into species diversity, pathogen suppression capabilities and carbon metabolism, HoloE2Plant designs SynComs that confer a variable degree of resistance to R. solani. This innovative approach paves the way for developing microbiomes optimised for plant protection and resilience. We believe that our microbial collections provide valuable resources for studying plant-microbiome evolution. Furthermore, future projects involving these microbial strains, as well as the technologies developed in HoloE2Plant to reconstruct SynComs, will support scientists in developing innovative biocontrol and biostimulant formulations.
Using SynComs in experimental evolution to track the coevolutionary processes between plants and microbiomes
How can SynComs be used to track coevolution between plants and microbiomes?
This is the overarching question addressed in HoloE2Plant. To study the real-time coevolution of hosts and their microbiomes, we leverage an exceptional model consisting of fast-cycling B. rapa plants. Professor Emeritus Paul H. Williams developed this smaller, rapidly growing variant of turnip at the Department of Plant Pathology, University of Wisconsin-Madison. These plants are ideal for studying plant evolution because they complete their life cycle (from seed to seed) in just five weeks, compared to over six months for natural (or cultivated) turnip populations.
In our project, we combine SynComs derived from natural B. rapa populations with fast-cycling plants to investigate the genetic bases of coevolution. Specifically, we aim to identify the genetic traits that are selected during coevolution and contribute to plant resilience against the fungal pathogen R. solani. To achieve this, we selected three SynComs with varying levels of microbial diversity, hypothesising that the degree of diversity influences the microbiome’s impact on plant health and production traits. These SynComs also include bacterial and fungal species with known (because tested experimentally in our laboratory) biocontrol activity against R. solani.
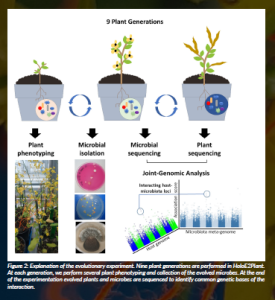
The experimental design involves coevolving these three SynComs with fast-cycling plants under controlled greenhouse conditions, both in the presence and absence of R. solani (Figure 2). At each plant generation, we measure several phenotypes related to disease resistance, plant fitness and growth. Simultaneously, we isolate the evolved microbial strains to build a collection for genomic characterisation. This experimental evolution approach is an exceptional tool that provides insights into the long-term effects of SynComs on plant health. Notably, it is rare for biocontrol formulations to be tested across multiple plant generations, which limits our understanding of their long-term efficacy. Thus, our experimental evolution approach offers a unique framework for dissecting the dynamics of plant-microbiome coevolution and paves the way for the development of plant probiotics. These probiotics would have demonstrated efficacy and safety over multiple generations, making them robust and reliable solutions for sustainable agriculture.
What comes next after experimental evolution?
Plant phenotypes, particularly those related to host fitness, collected over multiple generations and under varying SynCom regimes, provide invaluable insights into the dynamics of holobionts. However, to definitively establish holobionts as units of selection in evolution, it is essential to track evolutionary changes in both the host and its microbiome. In HoloE2Plant, we will address this by comparing the genomic features of ancestral and evolved plant populations alongside their corresponding SynCom populations. This will allow us to identify the genetic bases underlying coevolution (Figure 2). To tackle this specific question, we will develop and apply novel methods for joint association studies across organisms with divergent phylogenetic relationships.
Summary
HoloE2Plant has established a unique, extensively characterised fungal and bacterial collection that serves as a foundation for studying coevolution between plants and microbes.
The methodologies developed for SynCom reconstruction are powerful tools for identifying microbial combinations with biocontrol and biostimulant activities. When combined with experimental evolution, these approaches enable the development of microbial formulations with proven, long-term efficacy. This innovative approach can radically accelerate the research for alternative formulations to replace pesticides.
By uncovering the genetic bases in plants and microbes that are shaped by coevolution and contribute to enhanced disease resistance and fitness, HoloE2Plant paves the way for designing high-performing plant breeds and SynCom combinations. These innovations will offer greater adaptability to emerging epidemics and environmental challenges.
We hope that experimental evolution, extensive microbial collections from natural habitats and the methods we are developing will contribute to the creation of new eco-friendly products for crop protection.
PROJECT NAME
HoloE2Plant
PROJECT SUMMARY
The evolutionary processes underlying interactions between hosts and their associated microbes remain a black box that biologists have long sought to illuminate. A major limitation has been the lack of knowledge about precise molecular events, which are now increasingly accessible through next-generation sequencing technologies. The ERC Starting Grant HoloE2Plant project leads the way in host-microbiome evolutionary studies, utilising fast-cycling B. rapa plants and synthetic microbial communities. By harnessing high-throughput sequencing, experimental coevolution and advanced modelling, it unravels plant-microbiome interactions under pathogen pressure across multiple plant generations. By validating the holobiont concept and identifying key genetic mechanisms, HoloE2Plant sets the stage for designing microbial consortia for biocontrol and biostimulation, advancing functional evolutionary science in service of agroecology.
PROJECT LEAD PROFILE
Dr Claudia Bartoli, an evolutionary biologist at INRAE’s LIPME Laboratory in Toulouse, specialises in microbial evolution and plant health. Her research integrates genomic modelling, coevolution experiments and association studies to uncover plant and microbial genes linked to crop health. Driven by agroecology, she aims to develop innovative, microbe-based strategies to prevent crop diseases sustainably.
PROJECT CONTACTS
Claudia Bartoli (Project Coordinator)
Email: claudia.bartoli-kautsky@inrae.fr
FUNDING
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 101039541.
Figures
Figure 1: Representation of: (i) Brassica rapa wild populations collected for microbial isolation (left panel) and (ii) fungal diversity obtained by culturomics (right panel).
Figure 2: Explanation of the evolutionary experiment. Nine plant generations are performed in HoloE2Plant. At each generation, we perform several plant phenotyping and collection of the evolved microbes. At the end of the experimentation evolved plants and microbes are sequenced to identify common genetic bases of the interaction.