How much of the inequality in wealth, health and family planning can we explain with the data we have?
The desire to understand, explain and predict human behaviour has been a longstanding and central goal of the social sciences. Since the 1980s, empirical social research has increasingly emphasised methodological rigour, aiming to enhance the credibility of causal claims through statistical modelling. This tradition focuses on estimating the causal effects of specific variables—what can be called an ‘effect of causes’ approach. The objective is to determine whether and to what extent a given social factor, such as parental education or neighbourhood context, causally influences outcomes like educational attainment, fertility or depression.
Limits of causal inference in social science
More recently, researchers have turned to a different yet complementary set of questions. To what extent are social outcomes predictable? How much of the variation in an outcome across individuals can we explain, given the available data? This predictive, variance-decomposition perspective can be described as a ‘cause of effects’ approach, which shifts the focus from individual causal mechanisms to a broader analysis of outcome variance. Instead of isolating the effect of one variable, this perspective aggregates as many explanatory variables as possible to capture the complexity of social outcomes. It also provides critical insights into the limitations and potential of social science theories by evaluating the aggregate explanatory power of known predictors.
Assessing how well statistical models explain or predict outcomes—such as educational attainment, fertility or depression—has profound implications. It contributes to theory building, informs intervention design and enhances scientific discovery. Yet despite the promise, much of the recent work in this area suffers from notable limitations. It often relies on opaque ‘black box’ algorithms, fails to account for confounding by non-social (e.g. genetic) factors, and delivers disappointingly low predictive accuracy. As a result, many studies fail to achieve their goal of providing interpretable, robust and generalisable insights into the drivers of social outcomes.
From prediction to explanation: lessons from genetics
One major oversight is that recent social science efforts have not fully incorporated the methodological advances made in genetics over the past two decades. Quantitative geneticists have developed transparent and replicable analytical pipelines that directly address similar questions about variance decomposition. In particular, they have focused on quantifying the proportion of individual differences in traits that can be attributed to genetic and environmental factors. According to a comprehensive meta-analysis of all published twin studies, on average, about 50% of individual differences across a wide range of traits are associated with genetic variation (heritability), while the remaining 50% are linked to (social) environmental factors and measurement error (environmentality).
Heritability is defined as the proportion of total variance in a trait that can be statistically attributed to genetic differences within a given population. For social scientists, this concept is analogous to the R-squared in a regression model, where the genome is the set of predictors and the trait is the outcome. More intuitively, heritability tells us how much of the variation between individuals in a population is due to their genetic differences. Importantly, high heritability does not imply determinism or immutability—environmental interventions can still play a transformative role.
Findings of substantial heritability have spurred major investments in genome-wide association studies (GWAS) designed to identify specific genetic variants associated with traits of interest. These efforts have led to important discoveries, but also to the realisation that only a fraction of the heritability estimated from twin studies could be accounted for by discovered genetic variants. This gap, termed the ‘missing heritability’ problem, sparked decades of intense methodological innovation. Researchers developed new statistical models to account for small effect sizes distributed across many genetic variants, non-linear effects (dominance), gene-gene interactions (epistasis), and gene-environment interactions. They also began addressing the role of rare variants and measurement error in explaining the gap.
The FINDME approach
Our FINDME (Finding Missing Environmentality) project applies these insights to the social sciences. Just as geneticists have asked where the ‘missing heritability’ lies, we ask: where is the ‘missing environmentality’? That is, how much of the variance in outcomes attributed to environmental factors in twin studies can we actually explain using observed non-genetic variables? To answer this, FINDME brings together high-dimensional modelling, transparent causal inference and a comparative framework across datasets and societies. Our first step is to quantify the gap between the expected explanatory power of environmental influences (as estimated from twin studies) and the variance explained by observed, measured non-genetic variables. We anticipate that the observed explanatory power will fall far short of expectations, highlighting the ‘missing environmentality.’ We then adapt and extend methodological approaches from genetic research to better capture this unexplained variance. Specifically, we model the following:
- Social dominance effects, defined as non-linear effects of environmental variables on outcomes.
- Social epistasis, or the interactions between multiple environmental variables.
- Gene-environment interactions in which the effect of a social factor depends on genetic context.
- Measurement error and omitted variable bias.
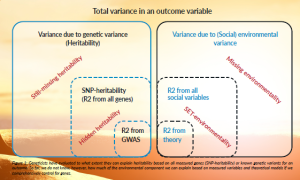
Figure 1 outlines this parallelisation. On the left, it shows the methodological toolkit used to decompose genetic variance, and on the right, our adaptation of these tools to environmental variables. One of the early lessons from the ‘missing heritability’ literature was that many small genetic effects are difficult to detect but collectively important. This has led to the development of polygenic scores and matrix approaches capable of modelling many small effects simultaneously. We propose doing the same for social variables. Instead of focusing on a small number of theoretically motivated predictors, we embrace high-dimensional modelling to uncover the cumulative and interactive effects of hundreds or thousands of social factors.
Modelling social complexity
For example, consider educational attainment. Traditional sociological models may include a handful of variables such as parental education, family income and neighbourhood characteristics. But datasets like the UK Biobank contain far more detailed measures: not just a binary indicator of neighbourhood deprivation, but a set of ten different indices capturing aspects such as economic hardship, exposure to crime and environmental hazards. When we consider 20 such variables, the number of possible interactions exceeds 1 000 000. Modelling this social complexity requires tools capable of handling such high-dimensional data, while still producing interpretable results.
Data, methods and comparative framework
FINDME systematically integrates these methods into the analysis of social outcomes. We adapt statistical techniques from genetics to jointly model genetic and environmental predictors, using extremely high-dimensional data. While classical twin models decompose variance into genetic and environmental components without specifying mechanisms, our approach allows us to specify, test and interpret complex models of gene-environment interplay. We do this using population-based datasets from Europe and the United States, including large-scale biobanks and national registers. Many of these datasets feature overlapping measures, which enables robust replication and cross-society comparison.
Key research questions
We focus on three critical outcomes: educational attainment, fertility and well-being. These traits have high societal relevance, are widely studied and are known to be influenced by both genetic and social factors. For each, we ask:
- How much of the variance can be explained by observed social variables?
- How much of this explanation is independent of genetic confounding?
- What is the contribution of gene-environment interactions?
- Do explanations generalise across social and geographic contexts?
Importantly, our aim is not only to increase predictive accuracy, but also to contribute to theory development. Are our current theories too simplistic? Do they underestimate the complexity of social life? Do findings vary systematically by cohort, region or institutional context? Do we see consistent patterns in how genetic and social factors interact?
Implications for theory and policy
By quantifying the relative contributions of different domains—genetic, environmental and their interactions—to the distribution of outcomes in populations, we aim to clarify where and how interventions may be most effective. Just as the discovery of polygenic scores has opened new avenues in personalised medicine, our work may open new avenues in targeted social policy and education reform. For instance, if social epistasis effects dominate in a given outcome, this suggests that policy interventions must be coordinated across domains (e.g. education and housing) to be effective. If gene-environment interactions are key, it implies that the same intervention may work differently depending on individual predispositions.
FINDME is both scientifically ambitious and practically urgent. As large-scale data collections continue to grow in scope and cost, it is critical to develop methods that can fully exploit their potential. While genetic compo are increasingly modelled with precision and sophistication, the modelling of social variables often remains rudimentary. This asymmetry limits our ability to evaluate theories, test interventions and understand social stratification.
Our project addresses this imbalance by bringing the methodological lessons of genetics into sociology. We do so not to reduce complex human behaviour to biological determinism, but to elevate the scientific rigour and explanatory capacity of social science. In doing so, we hope to shift the conversation from a dichotomy of nature versus nurture to a more nuanced and data-rich understanding of how both domains interact. We envision a future in which social science explanations are not only theoretically compelling but also empirically powerful—able to explain, predict and ultimately inform interventions that reduce inequality and improve well-being.
In sum, FINDME introduces a new paradigm for explanatory social science. It quantifies what we know, identifies what we don’t and builds a methodological bridge between genetics and sociology to uncover the missing pieces. With this work, we aim to advance not only our understanding of social outcomes but also the foundations of a more integrated and effective social science.
FINDME
(Finding Missing Environmentality)
Project summary
This project will evaluate to what degree we can explain and predict life course outcomes, such as fertility, education or well-being, based on social science and genetic data.
Project lead profile
Felix Tropf is the PI of the project. His research focuses on questions in social demography and quantitative genetics. His contributions to the field were recognised with the European Demography Award for best PhD Thesis.
Project contacts
Felix Tropf
Email: fctropf@gmail.com
Funding disclaimer
This project has been funded by UK Research and Innovation (UKRI) under the European Union’s Horizon Europe Guarantee programme – grant agreement number EP/Y023080/1.
Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or UKRI. Neither the European Union nor the granting authority can be held responsible for them.
Figure legends
Figure 1: Geneticists have evaluated to what extent they can explain heritability based on all measured genes (SNP-heritability) or known genetic variants for an outcome. So far, we do not know, however, how much of the environmental component we can explain based on measured variables and theoretical models if we comprehensively control for genes.